In my last article, we discussed Hydra [1], a tool for managing multiple experiments and configurations, which is particularly useful for cases of complex sets of experiments, such as the ones that happen in my daily work as a research scientist [2].
On the other hand, a couple of months ago, I presented MLFlow [3], a tool to track our machine learning experiments [4]. This tool provides a nice interface so we can easily compare different experiments, see them in tabular form, and have different kinds of interactive plots.
Both of these tools have been essential for me when working on experiments for academic papers and also while doing research for industry. Over the years, I have developed a framework for experimentation with them based on a few core ideas and how the combination of these ideas and these two frameworks helps me develop and prove hypotheses.
In this article, I'll introduce you to my experimentation framework both in terms of how I use it and in what instances it helps me deal with my job.
Tags in MLFlow
One feature we didn't discuss in my introductory post to MLFlow is the tags. The tagging system of MLFlow is useful for setting arbitrary tags [5]. These tags can be used to group different experiments and filter them via the search engine available in the Tracking UI of MLFlow. The code to set a tag is quite simple:
import mlflow
with mlflow.start_run():
mlflow.set_tag("tag_key", "tag_value")This snippet doesn't do much, but it will create a new experiment run in the database that has the tag "tag_key" associated with the "tag_value". The key can be any alphanumeric string, but it can also contain underscores (_), dashes (-), periods (.), spaces ( ), and slashes (/).
Let's remember that for MLFlow, a run is a single instance where the experiment is run using a specific set of parameters and records a set of metrics, while an MLFlow "experiment" is a way to group a set of MLFlow runs. You can also set a tag for a single experiment like so:
import mlflow
mlflow.set_experiment("experiment_name")
mlflow.set_experiment_tag("experiment_tag_key", "experiment_tag_value")Modifying the Description of Experiments and Runs
Within the possible tags, both for experiments and runs, there are a couple of system tags that serve different functionalities. In particular, the tag mlflow.note.content is a tag that modifies the "notes" or description of the experiment or run, accepting text in Markdown format [6]. This tag is essential to the experimentation framework we'll be seeing in this blog article:
import mlflow
from sklearn.datasets import load_iris
from sklearn.metrics import accuracy_score, classification_report
from sklearn.model_selection import train_test_split
from sklearn.svm import SVC
data = load_iris()
X_train, X_test, y_train, y_test = train_test_split(
data['data'], data['target'],
train_size=0.8, random_state=42
)
kernels = ["linear", "rbf", "poly"]
mlflow.set_experiment("SVM-Iris")
mlflow.set_experiment_tag(
"mlflow.note.content",
"# SVM over Iris\n\nExperiment of different SVM kernels over Iris Dataset."
)
for kernel in kernels:
with mlflow.start_run(run_name=f"kernel={kernel}"):
mlflow.log_param("kernel", kernel)
clf = SVC(kernel=kernel).fit(X_train, y_train)
y_pred = clf.predict(X_test)
mlflow.log_metric("accuracy", accuracy_score(y_test, y_pred))
mlflow.set_tag(
"mlflow.note.content",
"# Classification Report\n\n"
f"```\n{classification_report(y_test, y_pred)}\n```"
)In the previous snippet, we run 3 experiments over the Iris Dataset [7]. The experiment runs use the support vector machine classifier model with 3 different kernels: linear, radial basis function (rbf), and polynomial (poly) over the same split of train and evaluation data. There isn't much content in the code itself, and the results aren't really useful since all three models finish with perfect results. The important part of the code is the set of system tags that change the description of both the experiment and each of the runs. If we start the MLFlow UI and see the experiment we have just created, we'll see the following:
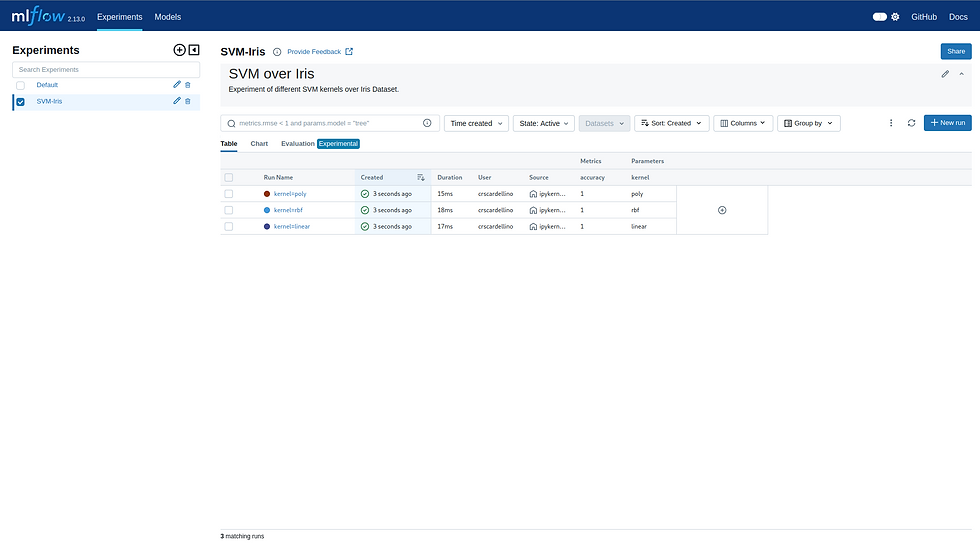
As you can see, over the table with the three runs, we can see the description of the experiment. Since we used the markdown tag for a heading in the first line of the description (i.e., we started the line with #), there is a title for the description, followed by the next line without formatting.
If we pick any run to see its full overview, we'll see that at the very beginning of the overview, where the description is, we have written directly the classification report of Scikit Learn [8]. We made use of the triple backquote in markdown (```) when writing this tag in order to show it with a monospace font that is easier to visualize.

The Experimentation Framework
In my daily job, the combination of Hydra and MLFlow can help me keep track of the different experiments I have to do. Even though these tools are very good when trying to find the best possible combination of parameters that renders the best result over the evaluation or development data, this is not the main reason I used them.
More often than not, especially when working in a more academic setting, where there's a high level of uncertainty, the simple objective of finding the best possible setup for the highest results is not enough. Good research publications shouldn't be concerned with having good performance on evaluation data but rather should understand why certain configurations render that performance.
Moreover, when working in academic research, I usually have to deal with multiple hypotheses that are wildly different and test things from the same experimentation that are very far apart from each other.
Using MLFlow for Hypothesis Testing
With this idea in mind, I started designing a framework to help me in this task. The main build of this framework is that each group of experiments tests a hypothesis. So, instead of grouping experiments runs by the dataset used to run them or by the type of model used, I define a hypothesis, and under that hypothesis, my runs try to test it to the fullest.
I leverage the MLFlow experiment name and the experiment description tag to clearly define what hypothesis I am testing within that set of experiments. This way, I can run several hypotheses that test different things without having to worry about what I was trying to prove with that set of experiments. An example of a hypothesis could be: "Hypothesis: Adding more layers in a multilayer perceptron produces overfitting".
Each run from a set of experiments defines the different configurations that serve to accept or reject the hypothesis I am testing (I know that technically, we never accept a hypothesis, so take the accept in here with a grain of salt, you know what I mean). To avoid cluttering, I only log the main parameters I have for each run, and so it is easier to visually differentiate which run is which, I either use some key values being tested as part of the name of the run or I add it to the description of that run.
With the artifact logging provided by MLFlow, I can either save the full set of parameters or the configuration of the model or, if I deem it useful, I can even log the pickled model directly as an artifact.
If I combine MLFlow with Hydra, the multiple configurations required to test a Hypothesis become easier to manage, as I can modify any configuration or even add new configurations directly from the CLI [9] without having to code them with argparse or even add them to a configuration file, although I prefer to set the configuration file with as much of the parameters I will be using as possible.
Using MLFlow for Reducing the Parameter Search Space
Another useful functionality I found in MLFlow is a tool that helps reduce the parameter search space. So, how does this work? Follow along as I try to unravel this.
When I am working with high uncertainty, for example, when I am building a new neural network architecture or when I am adapting some architecture to a new domain, the search space can become quite large to explore as a whole. Generally, I start by setting up everything to the standard values for all hyperparameters, which are usually the default values in whatever framework or library I am using for the task. This generally gives me a good idea of whether or not the model I am developing is useful for the task at hand, i.e., if the default values give a very poor performance, that is a good indication the model might not be suitable for the problem.
After we have cleared that the model we chose is suitable for the problem, we need to explore some hyperparameters to improve on the final result, i.e., we have to do hyperparameter optimization. If you read my previous article on strategies for machine learning in the industry [10], you know that we should go with a random search first to reduce the search space of parameters. If we go with a grid search first and we have a large search space, the task can become completely intractable in the number of required experiments.
In machine learning, and especially in deep learning, there is an important component of randomness. The seed that is used for setting up the models can be a game changer under certain configurations. However, when doing a random search, I found that it is better to have that fixed and check the other combinations of hyperparameters.
MLFlow provides scatterplots, and these are very good for visually identifying those hyperparameters that made a change. When you run a couple of experiments with random hyperparameters, it is generally noticeable that those hyperparameters have a great deal of impact on the final result. Once I identify those hyperparameters, I can start making hypotheses about the type of changes such hyperparameters require for the final model to improve. For example, suppose I notice that using more layers in a neural network decreases the overall result regardless of other hyperparameters. In that case, I can focus on testing the hypothesis that "more layers cause overfitting". For testing the hypothesis, I made use of random seeds that gave me a better overall idea of whether or not the results were a fluke.
An Example Experimentation Application
To exemplify the application of this experimentation framework, I have created a repository on GitHub [11]:
$ git clone https://github.com/crscardellino/hydra-mlflow
$ cd hydra-mlflowThe repository has the following structure:
.
├── data
│ └── wines-data.csv
├── LICENSE
├── mlflow_hydra
│ ├── conf
│ │ └── config.yaml
│ ├── experiment.py
│ ├── __init__.py
│ ├── model.py
│ └── utils.py
├── NOTICE
├── README.md
└── requirements.txtThe example application for experimentation is in the mlflow_hydra module. The data for this application comes from the wines dataset [12]. To set up the application, simply create a virtual environment and install the requirements:
$ python -m venv venv
$ source ./venv/bin/activate
(venv) $ pip install -r requirements.txtThe Configuration File
There are two main sections in the configuration file:
The input section defines the experiment properties. Particularly, besides the path to the data file, I define things such as the hypothesis (which is the name of the MLFlow experiment), the description of such hypothesis, and the name of the MLFlow run that I'll use to highlight the most important information (i.e., the hyperparameters) to be easily accessible.
In the train section, I define the implementation details to use in the experiment. Properties such as the hyperparameters to test in the different runs of the experiment will facilitate in accepting or rejecting the hypothesis for such an experiment.
The configuration file is the following:
input:
data_file: ???
run_name: ???
experiment_name: Hypothesis 1
experiment_description: '**Hypothesis 1:** More layers increase the overfitting'
train:
test_evaluation: false
feature_scaling: true
batch_size: 16
epochs: 10
early_stop: 3
model:
layers:
- 64
learning_rate: 0.001
l2_lambda: 1.0e-05
activation: ${eval:torch.nn.ReLU}
random_seed: 42The Experiment Application
The experiment application, defined in the experiment.py module, has a main function that will be in charge of doing the initial setup of MLFlow (i.e., define the experiment name, description, logging hyperparameters, etc.). It also has a function called run_experiment, which will be in charge of running the Pytorch Lightning [13] model that also binds to the MLFlow logger [14] to record information about the model training. All the code is documented, so please check them out for more information.
Run a Set of Experiments for a Hypothesis
To run a set of experiments with different hyperparameters, given a hypothesis, to accept it or reject it (again, accept is a strong word here, we use it for simplification's sake). You can use whatever method you prefer, I tend to use BASH scripts:
LAYERS="[] [64] [64,64] [64,64,64]"
SEEDS="0 42 1337"
for layers in $LAYERS
do
for seed in $SEEDS
do
python -m mlflow_hydra.experiment \
input.data_file=./data/wines-data.csv \
input.run_name=\"layers:$layers\" \
train.model.layers=$layers \
train.random_seed=$seed
done
doneAs you can see, I test with three different random seeds for this, since it gives me more confidence concerning the final results.
Once the experiment is run, we can see the final experiments and compare them with a table comparison:

Where we have access to the full data. We can also plot them, in this case only those that are of the same random seed, to have a better visualization of them:

Finally, my favorite type of comparison when testing a hypothesis is the scatterplot, where I compare the relevant metric (in this case, the validation loss) against whatever hyperparameter I'm focusing on (in this case, the number of layers). As we can see, it's clear that the hypothesis: "More layers increase overfitting" doesn't hold up to the results, which show the contrary, and thus we can reject such hypothesis:

Concluding Remarks
In this article, I presented one experimentation framework I find useful to work with. Of course, it can be improved, that's why I wanted to openly share it for other people to use it if they think it fits in their daily jobs. The main tools I use are MLFlow and Hydra, alongside a pattern to follow. I hope you can make use of it, too, and feel free to accommodate it to your needs.
References
[1] Hydra. A framework for elegantly configuring complex applications. https://hydra.cc/
[2] Cardellino, C. 2024. Tracking Multiple Experiments with Hydra. Transcendent AI. https://www.transcendent-ai.com/post/tracking-multiple-experiments-with-hydra
[3] MLFlow: ML and GenAI made simple. https://mlflow.org/
[4] Cardellino, C. 2024. Keeping Track of Experiments with MLFlow. Transcendent AI. https://www.transcendent-ai.com/post/keeping-track-of-experiments-with-mlflow
[5] MLFlow Documentation. Adding Tags to Runs. https://mlflow.org/docs/latest/tracking/tracking-api.html#adding-tags-to-runs
[6] Markdown Guide. https://www.markdownguide.org/
[7] Fisher, R.A., 1936. The use of multiple measurements in taxonomic problems. Annals of eugenics, 7(2), pp.179-188.
[8] Scikit Learn. Classification Report. https://scikit-learn.org/stable/modules/generated/sklearn.metrics.classification_report.html
[9] Hydra Documentation. A simple command-line application. https://hydra.cc/docs/tutorials/basic/your_first_app/simple_cli/
[10] Cardellino, C. 2024. Practical Machine Learning for Industry. Transcendent AI. https://www.transcendent-ai.com/post/practical-machine-learning-for-industry
[11] Cardellino, C. 2024. MLFlow + Hydra: A Framework for Experimentation with Python. https://github.com/crscardellino/hydra-mlflow
[12] Aeberhard,Stefan and Forina,M.. (1991). Wine. UCI Machine Learning Repository. https://doi.org/10.24432/C5PC7J.
[13] Lightning AI. PyTorch Lightning Documentation. https://lightning.ai/docs/pytorch/stable/
[14] Lightning AI. Pytorch Lightning Documentation. MLFlow Logger. https://lightning.ai/docs/pytorch/stable/api/lightning.pytorch.loggers.mlflow.html

Comments